Case Studies
Engineering Selectivity in a Challenging Target
Unique pharmacophore technology leads to rapid creation of novel, selective hits for DNMT1
Challenge: Off target effects limit efficacy of drugs targeting DNMT1
DNMT1 is an enzyme linked to multiple cancers. It acts on the same natural substrate as its close relatives, DNMT3a and DNMT3b. This overlap makes it difficult to design drugs that interact specifically with DNMT1 without also affecting the others.
Current drugs avoid this challenge by avoiding the enzyme’s active site and instead bind to the DNA substrate. While this indirect approach can be effective, it causes unwanted side effects.
DNMT1 Transition State - Methyl Transfer from cofactor to DNA
Solution: Transition state based DNMT1 inhibitors
Even very similar enzymes accelerate reactions in subtly different ways. We used our platform to discover these variations and design drugs that exploit them to bind to only the intended target. The key to this is identifying the turning point in the reaction - the “Transition State”.
How we did it
In only 2 months, our multidisciplinary team progressed from the crystal structure of DNMT1 to designing novel inhibitors:
Our computational scientists deployed automated simulation workflows to capture the enzyme’s reaction dynamics and pinpoint the transition state*.
Our proprietary Quantum Pharmacophore technology, we extracted the key shape, electrostatic, and interaction features that define the transition state and distinguish DNMT1 from its close relatives.
The drug design team fed the Quantum Pharmacophore into our generative AI engine to produce candidate molecules.
Through a combination of computational scoring, structural visualisation, and expert insight, we prioritised compounds for synthesis and experimental validation in the wet lab.
*We used so-called QMMM simulations which combine a quantum region to describe the chemistry and a less accurate but cheaper classical model for the wider enzyme.
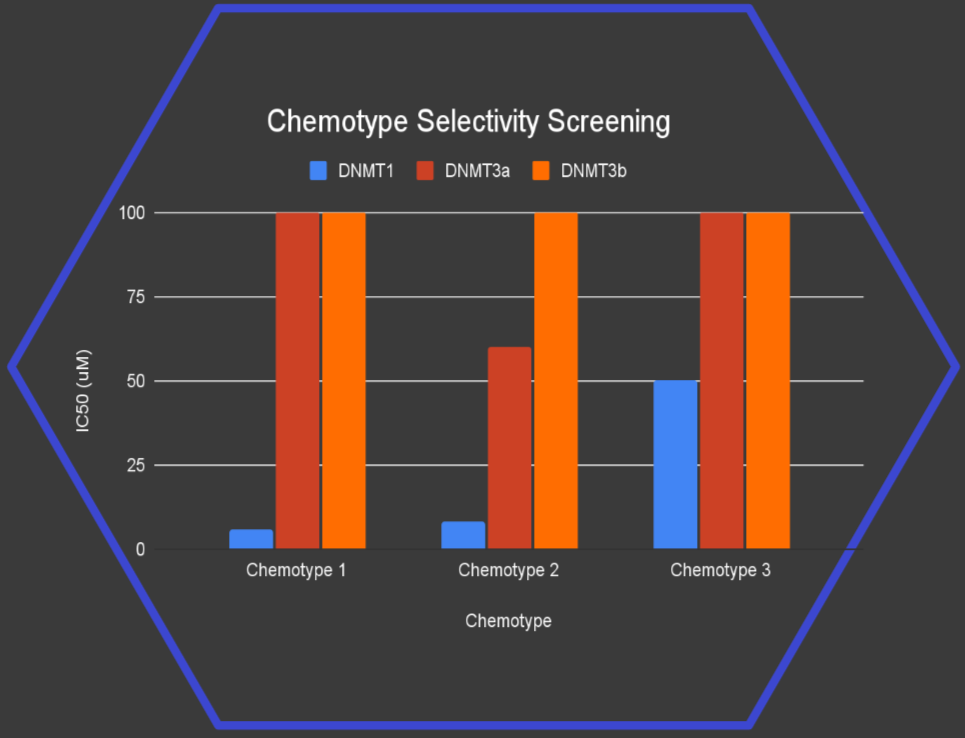
Result: Selective DNMT1 inhibitors
We made and tested just 40 compounds, yet found 3 novel chemotypes* active against DNMT1 with strong selectivity over DNMT3a and DNMT3b.
Notably, 2 of these chemotypes match the potency of well-established hit series reported in the scientific literature, marking a significant step forward in the search for safer, more targeted therapies.
* A chemotype is a class of molecules with related structures
Pinpointing Selectivity Through Quantum Energy Profiling
Quantum interaction analysis decodes selectivity beyond sequence differences in SHP1/2 phosphatases
Challenge: Unexplained selectivity between closely related phosphatases
In 2024, researchers publised a natural product derived series of inhibitors which were found to exhibit selectivity between phosphatases in the PTP subfamily. Most of the observed differences in binding strength could be explained by changes in amino acid sequence between the different phosphatases. However, between two of the most closely related - SHP1 and SHP2 - no differences close to the active site were present but the inhibitor bound ten times strongly to SHP2. Understanding this selectivity is key to designing inhibitors without unfavourable off target effects.
Solution: Quantum Interaction Analysis
We employed both quantum and classical methods to explain the difference between the two phosphatases. Using the Quantum Interaction component of our Quantum Lens platform we were able to provide quantitative results that explained the selectivity. This method was based on Fragment Molecular Orbital and pair interaction energy decomposition analysis (PIEDA) techniques which allowed us to:
Decompose both attractive & negative components
Capture charge transfer
Look beyond active site
Result: Peripheral residue energies drive selectivity
Our Quantum Lens analysis revealed that more than 50% of the binding contributions for the inhibitor came from peripheral residues (blue in the picture) beyond the active site (pink).
The differences in these residues were not picked up in the original publication but our modelling shows that comparatively unfavourable interactions far from the active site could explain the difference in binding between SHP1 and SHP2. In particular a comparison of the peripheral residue energies showed a lysine (LYS) contributing a large repulsive energy only in the SHP1 complex. This identification of an unintuitive residue interaction can enable enhanced optimisation of mildly selective compounds.
Pipeline
Kuano’s innovative quantum approach to drug discovery enables unique targets to be explored to provide innovative therapies.
“Apart from checkpoint inhibitors for a few, there have been few advances in colorectal cancer in the last 20 years. Kuano's novel approach exploiting quantum mechanics has unlocked a unique target that permits an exciting biologically effective novel therapy for a much larger proportion of cancer sufferers.”
John Bridgewater, Clinical Professor, UCL